Ruminal Methanogen Community Changes in Response to the Rumen Development and the Addition of Rhubarb
As the environmental awareness rises dramatically all over the world, increasing concern has been placed on the GHG (greenhouse gas) emission. Amongst the various GHGs, methane (CH4) is around 20-30 times more damaging than CO2 in global warming. Ruminants (e.g., cattle, sheep, and goats), in fact, are the major producers of methane, generally taking up 15-25% of global emission of methane. More specifically, it is the ruminal methanogens that produce methane during rumen fermentation in ruminants. In addition, methane produced by ruminal methanogens causes the loss of about 2–12% of the gross energy intake of the host.
It is held by many researchers that different types of microflora within the rumen are tightly bound up with different feed efficiencies and production performances of ruminants. Some previous investigations have further inferred that it would be practicable to modulate the rumen microbial community at the early period of rumen development, and early dietary regulations of animal have a greater and more lasting effect than those occurring later in life. Similarly, it could be possible to reduce methane production of ruminant husbandry in the long term by early manipulations on methanogens in young ruminants. Therefore, it is of great significance to explore the evolution of methanogen colonization as the rumen develops, and seek strategies in shaping the methanogenic community.
Recently, an international research performed by the Institute of Subtropical Agriculture,Chinese Academy of Sciences(ISA) and the Lethbridge Research Centre (Agriculture and Agri-Food Canada) together has clarified the changes of ruminal methanogen community during rumen development (from 1 d to 60 d after birth) in Xiangdong black goats (a local breed of goat in Changsha), and how rhubarb supplementation affected the methanogenic community.
“We used RNA isolated from the rumen samples instead of DNA, since RNA can help us gain insights into the real-time activities and metabolic potential of ruminal methanogens.” Said Zuo Wang, a doctoral student primarily conducting this study. RT-qPCR and 16S rRNA amplicon sequencing both based on RNA were performed to investigate the methanogenic population in the rumen of black goats, and the results indicated that Methanobrevibacter, Candidatus Methanomethylophilus, and Methanosphaera were the top three genera across four different fractions. The discrepancies in the distribution of methanogens across four fractions, and various fluctuations in abundances among four fractions according to age were noted, respectively. The addition of rhubarb significantly reduced the abundances of Methanimicrococcus spp. in four fractions on 50 d, but did not alter the methanogen community composition on 60 d.
In summary, this is the first study that uses RNA based samples to investigate the development of the potentially active methanogens in the rumen. It contributes to the knowledge of the development of the rumen methanogen community and relevant modulation, and mitigation of methane production during rumen development.
This research received funds from the National Natural Science Foundation of China (Grant No. 31561143009, 31320103917) and Hunan Provincial Creation Development Project (Grant No. 2013TF3006). This study was also supported by the Open Foundation of Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences (Grant No. ISA2016301), Youth Innovation Promotion Association CAS, and the MOE-AAFC PhD Research Program of the China Scholarship Council.
The present study entitled “Investigation and manipulation of metabolically active methanogen community composition during rumen development in black goats” has been published online by Scientific Reports of the Nature Publishing Group. (https://www.nature.com/articles/s41598-017-00500-5) 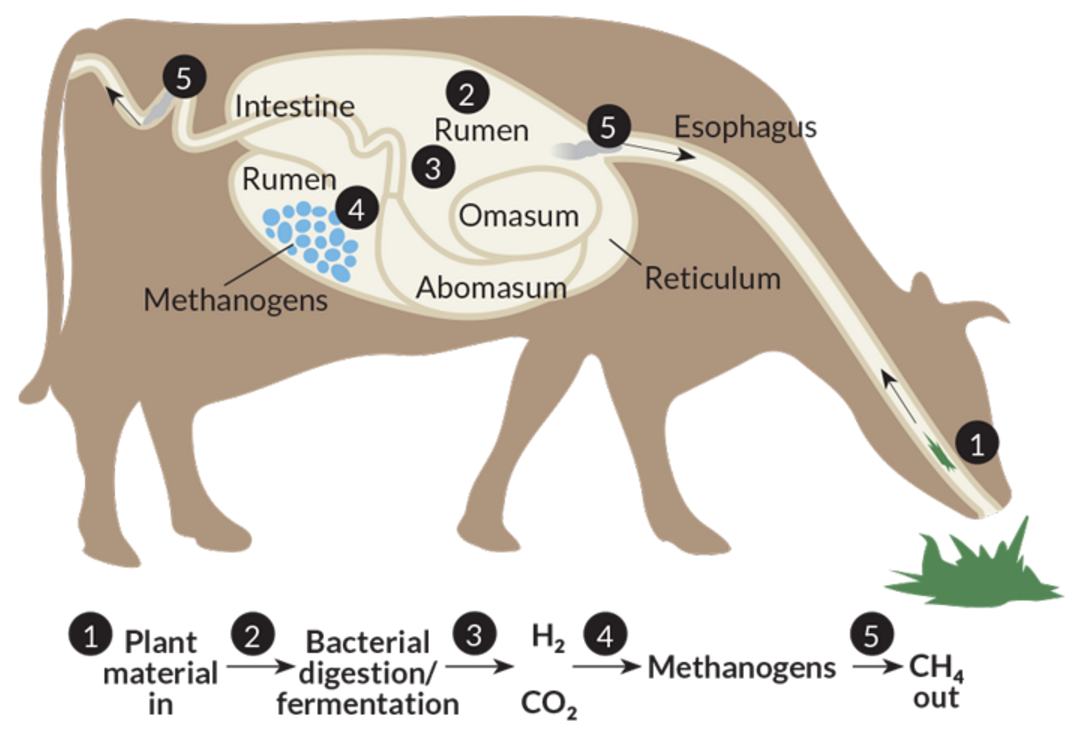
Processes of the methane production in ruminants
(https://www.sciencenews.org/sites/default/files/sn-2015/112815_cow_digestion_730_free.png)
Contact: TAN Zhiliang
E-mail: zltan@isa.ac.cn
Institute of Subtropical Agriculture, Chinese Academy of Sciences
Download attachments: